|
|
SYSTAT : SIGMAPLOT : TableCurve2D : TableCurve3D : PeakFIT : AutoSignal |
PRODUCT USES |
|---|
AGAR PLATE COLONY COUNTING AND SIZE DETERMINATION
The objective is to use image analysis to count and determine the size of bacterial colonies found on an agar plate. SigmaScan Pro is used to automate colony counting and measurement. The procedure described here, or modifications of it, can be used for simple colony counting or more complex analyses of colony data. It has application in the areas of immunology, bacteriology, and microbiology.
The agar plate image was captured using an inexpensive solid-state CCD camera and a standard off-the-shelf frame grabber board. The agar plate was back lit using a standard light box. The image is shown in Figure 1.
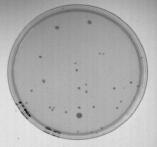
Figure 1: Original image of agar plate
Click to View Larger Image
The image was spatially calibrated using the Image, Calibrate, Distance and Area menu option. A 2-Point Rescaling calibration was performed using millimeter units. Colony areas will then be reported in square millimeters.
Uneven lighting over the image was corrected by using a pseudo-clearfield operation. This was done by selecting Clearfield, Generate Pseudo-Clearfield from the Image menu. The pseudo-clearfield was calculated using a rectangular grid with 8 x and y divisions. Figure 2 shows the pseudo-clearfield that is used to correct for the uneven illumination of the plate. The plate edge is visible in the clearfield image. This is an artifact of the "pseudo" correction process that for this application won’t affect the measurements. It would cause errors if intensity measurements were being made.

Figure 2: Pseudo clearfield used to correct for uneven background illumination
Click to View Larger Image
Select Clearfield, Perform Equalization… from the Image menu to correct the original image. The result is shown in Figure 3.
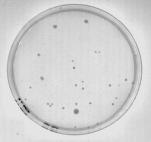
Figure 3: Image corrected for unequal background lighting
Click to View Larger Image
Increasing the contrast between colonies and the surrounding region will help identify the colonies by thresholding. Image contrast is improved by performing a Histogram Stretch from the Image, Intensity menu. This procedure measures the gray levels in the image. The user then "stretches" the range of gray levels with significant magnitude over the entire 255 level intensity range. In this case moving the Old Start line with the mouse to an intensity of 64 will eliminate the effect of the insignificant dark gray levels and improve the contrast. The results are shown in Figure 4.
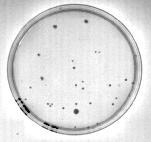
Figure 4: Contrast enhancement by "histogram stretching".
Click to View Larger Image
The colonies can be identified by thresholding the intensity level to fill in the darkest objects. This is done by selecting Threshold, Intensity Threshold from the Image menu. A dark gray intensity range of about 60 to 160 was used to identify the objects shown in the green overlay plane in Figure 5.
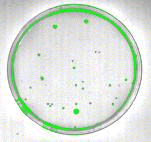
Figure 5: Use of an intensity threshold to identify dark objects.
Click to View Larger Image
The objects that are identified include portions of the edge of the plate. These objects can be removed by using image overlay layer math to intersect the bacterial colony objects with an overlay plane consisting of the interior of the agar plate. We will create the latter overlay by filling light pixels in the interior of the plate. Move your cursor radially from the interior of the plate toward the edge and observe the pixel intensity values. As you approach the plate edge the intensity decreases in the darker pixels. In Figure 5 the plate edge has an approximate intensity of 180. Select the Fill tab In the Measurement, Settings… dialog and select the Manual Threshold method. Set the Level to be 180 and the option to select objects that are lighter than this level. Select the Fill Measurement mode (paint bucket icon) and left click in the interior of the plate to fill it. You may need to set the source overlay to red in the Measurements, Settings, Overlays dialog. There are "holes" in the red overlay plane that were not filled since they contain dark pixels from the bacterial colonies. To fill them select Image, Overlay Filters and select the Fill Holes option. Let both the source and destination overlays be red. The resulting image is shown in Figure 6. The red circular overlay plane contains the green bacterial colonies.

Figure 6: A red overlay containing the green colonies to be measured.
Click to View Larger Image
The overlay math feature is used to identify the intersection of the red and green overlay planes. From the Image menu select Overlay Math and specify red and green to be the source layers and blue to be the destination layer. Then AND the two layers to obtain the intersection. The resulting image is shown in Figure 7.
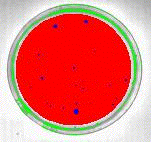
Figure 7: Identification of bacterial colonies (blue) by intersecting the red and green overlay layers.
Click to View Larger Image
The blue pixels overlay the bacterial colonies that can now be measured. Select the blue overlay plane as the source overlay from the Overlays tab in the Measurement Settings dialog. Select Perimeter, Area, Shape Factor, Compactness and Number of Pixels from the Measurements tab in the Measurements Settings dialog. Then measure the bacterial colonies using Measure Objects from the Measurements menu. The object numbers can be placed on the image as shown in Figure 8 and the measured quantities placed into the worksheet, Figure 9.
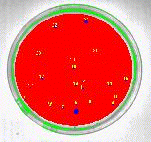
Figure 8: Colony label numbers displayed after object measurements.
Click to View Larger Image
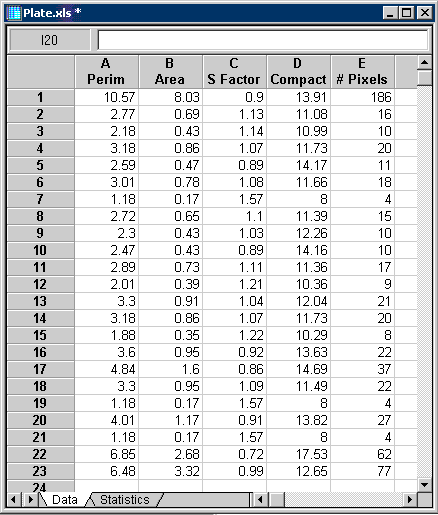
Figure 9: Bacterial colony measurement results.
1. Colin T. Sim, Labtronics, Inc